Copy
API_URL = "https://api.bluesight.ai"
GSD = 1.0
CHIP_SIZE = 224
COLLECTION = "naip"
BANDS = ["red", "green", "blue", "nir"]
DATA_DIR = "./data"
N_TRAIN_SAMPLES = 1000
N_TEST_SAMPLES = 100
RANDOM_SEED = 42
HEADERS = {"Content-Type": "application/json"}
Copy
!pip install awscli numpy scikit-learn gdown requests-toolbelt h5py rasterio
Loading Data
Copy
!aws s3 cp s3://us-west-2.opendata.source.coop/agentmorris/lila-wildlife/lcmcvpr2019/cvpr_chesapeake_landcover/ny_1m_2013_extended-debuffered-train_tiles/ data/train/ --recursive --no-sign-request --region us-west-2 --exclude "*" --include "*_lc.tif" --include "*_naip-new.tif"
!aws s3 cp s3://us-west-2.opendata.source.coop/agentmorris/lila-wildlife/lcmcvpr2019/cvpr_chesapeake_landcover/ny_1m_2013_extended-debuffered-val_tiles/ data/val/ --recursive --no-sign-request --region us-west-2 --exclude "*" --include "*_lc.tif" --include "*_naip-new.tif"
Preparing Data
https://github.com/Clay-foundation/model/blob/main/finetune/segment/preprocess_data.pyCopy
import os
import re
from pathlib import Path
import numpy as np
import rasterio as rio
from tqdm import tqdm
DATA_DIR = Path(DATA_DIR)
OUTPUT_DIR = DATA_DIR / "output"
Copy
def read_and_chip(file_path, chip_size, output_dir):
"""
Reads a GeoTIFF file, creates chips of specified size, and saves them as
numpy arrays.
Args:
file_path (str or Path): Path to the GeoTIFF file.
chip_size (int): Size of the square chips.
output_dir (str or Path): Directory to save the chips.
"""
os.makedirs(output_dir, exist_ok=True)
with rio.open(file_path) as src:
data = src.read()
n_chips_x = src.width // chip_size
n_chips_y = src.height // chip_size
chip_number = 0
for i in range(n_chips_x):
for j in range(n_chips_y):
x1, y1 = i * chip_size, j * chip_size
x2, y2 = x1 + chip_size, y1 + chip_size
chip = data[:, y1:y2, x1:x2]
chip_path = os.path.join(
output_dir,
f"{Path(file_path).stem}_chip_{chip_number}.npy",
)
np.save(chip_path, chip)
chip_number += 1
def process_files(file_paths, output_dir, chip_size):
"""
Processes a list of files, creating chips and saving them.
Args:
file_paths (list of Path): List of paths to the GeoTIFF files.
output_dir (str or Path): Directory to save the chips.
chip_size (int): Size of the square chips.
"""
for file_path in tqdm(file_paths):
read_and_chip(file_path, chip_size, output_dir)
train_image_paths = list((DATA_DIR / "train").glob("*_naip-new.tif"))
train_label_paths = list((DATA_DIR / "train").glob("*_lc.tif"))
val_image_paths = list((DATA_DIR / "val").glob("*_naip-new.tif"))
val_label_paths = list((DATA_DIR / "val").glob("*_lc.tif"))
process_files(train_image_paths, OUTPUT_DIR / "train/chips", CHIP_SIZE)
process_files(train_label_paths, OUTPUT_DIR / "train/labels", CHIP_SIZE)
process_files(val_image_paths, OUTPUT_DIR / "val/chips", CHIP_SIZE)
process_files(val_label_paths, OUTPUT_DIR / "val/labels", CHIP_SIZE)
Copy
100%|█████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 100/100 [13:09<00:00, 7.90s/it]
100%|█████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 100/100 [02:48<00:00, 1.68s/it]
100%|█████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 5/5 [00:43<00:00, 8.78s/it]
100%|█████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 5/5 [00:08<00:00, 1.60s/it]
Copy
train_chip_names = [chip_path.name for chip_path in list((OUTPUT_DIR / "train/chips").glob("*.npy"))]
train_label_names = [re.sub("_naip-new_", "_lc_", chip) for chip in train_chip_names]
val_chip_names = [chip_path.name for chip_path in list((OUTPUT_DIR / "val/chips").glob("*.npy"))]
val_label_names = [re.sub("_naip-new_", "_lc_", chip) for chip in val_chip_names]
Copy
from pathlib import Path
import h5py
import numpy as np
from tqdm import tqdm
num_bands = len(BANDS)
dt = np.dtype(
[
("bands", f"S10", (num_bands,)), # Adjust '10' if your band names are longer
("gsd", "float32"),
("pixels", "float32", (num_bands, CHIP_SIZE, CHIP_SIZE)),
("platform", "S20"), # Adjust '20' if your platform names are longer
("point", "float32", (2,)),
("timestamp", "int64"),
("label", "int64", (CHIP_SIZE, CHIP_SIZE)),
]
)
label_mapping = {1: 0, 2: 1, 3: 2, 4: 3, 5: 4, 6: 5, 15: 6}
output_file = "land_segmentation_train_data.h5"
with h5py.File(output_file, "w") as f:
dataset = f.create_dataset(
"data", (N_TRAIN_SAMPLES,), dtype=dt, compression="gzip", compression_opts=1, chunks=(min(1, N_TRAIN_SAMPLES),)
)
for i, chip_name in enumerate(tqdm(train_chip_names[:N_TRAIN_SAMPLES])):
label_name = re.sub("_naip-new_", "_lc_", chip_name)
dataset[i] = (
[band.encode("ascii", "ignore") for band in BANDS],
GSD,
np.load(OUTPUT_DIR / "train/chips" / chip_name).astype("float32"),
COLLECTION.encode("ascii", "ignore"),
[0, 0],
0,
np.vectorize(label_mapping.get)(np.load(OUTPUT_DIR / "train/labels" / label_name).squeeze()).astype("int64")
)
Copy
100%|███████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 1000/1000 [00:41<00:00, 23.96it/s]
Copy
import numpy as np
import matplotlib.pyplot as plt
def visualize_image_mask_pair(image, true_mask, pred_mask=None, title="Image and Mask Comparison"):
color_map = {
0: (0, 0, 255), # Deep Blue for water
1: (34, 139, 34), # Forest Green for tree canopy / forest
2: (154, 205, 50), # Yellow Green for low vegetation / field
3: (210, 180, 140), # Tan for barren land
4: (169, 169, 169), # Dark Gray for impervious (other)
5: (105, 105, 105), # Dim Gray for impervious (road)
6: (255, 255, 255), # White for no data
}
def create_colored_mask(mask):
colored_mask = np.zeros((*mask.shape, 3), dtype=np.uint8)
for i in range(mask.shape[0]):
for j in range(mask.shape[1]):
colored_mask[i, j] = color_map[mask[i, j]]
return colored_mask
true_colored_mask = create_colored_mask(true_mask)
n_cols = 3 if pred_mask is not None else 2
fig_width = 6 * n_cols
fig_height = 6
fig, axes = plt.subplots(1, n_cols, figsize=(fig_width, fig_height))
if n_cols == 2:
axes = [axes[0], axes[1]]
axes[0].imshow(image)
axes[0].set_title("Original Image", fontsize=12)
axes[0].axis('off')
axes[1].imshow(true_colored_mask)
axes[1].set_title("Ground Truth Mask", fontsize=12)
axes[1].axis('off')
if pred_mask is not None:
pred_colored_mask = create_colored_mask(pred_mask)
axes[2].imshow(pred_colored_mask)
axes[2].set_title("Predicted Mask", fontsize=12)
axes[2].axis('off')
plt.suptitle(title, fontsize=14)
plt.tight_layout()
plt.show()
with h5py.File(output_file, "r") as f:
dataset = f["data"]
random_indices = np.random.choice(len(dataset), 5, replace=False)
for i, idx in enumerate(random_indices):
image = dataset[idx]['pixels'].astype("int64").transpose(1, 2, 0)
true_mask = dataset[idx]['label']
if image.shape[2] > 3:
image = image[:, :, :3]
visualize_image_mask_pair(image=image, true_mask=true_mask, title=f"Random Train Sample {i+1}")
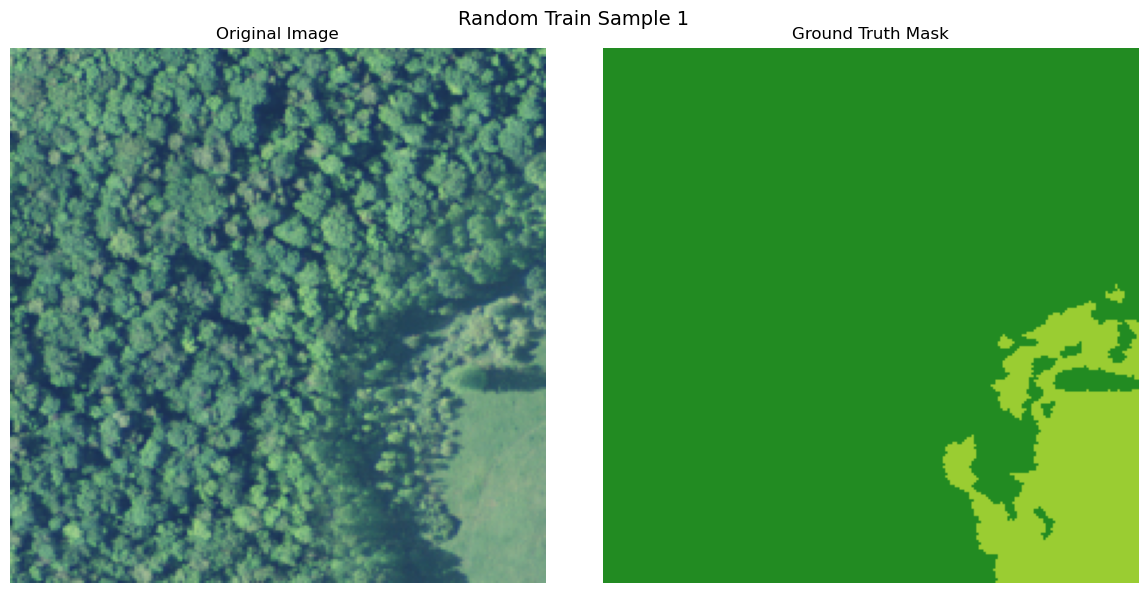
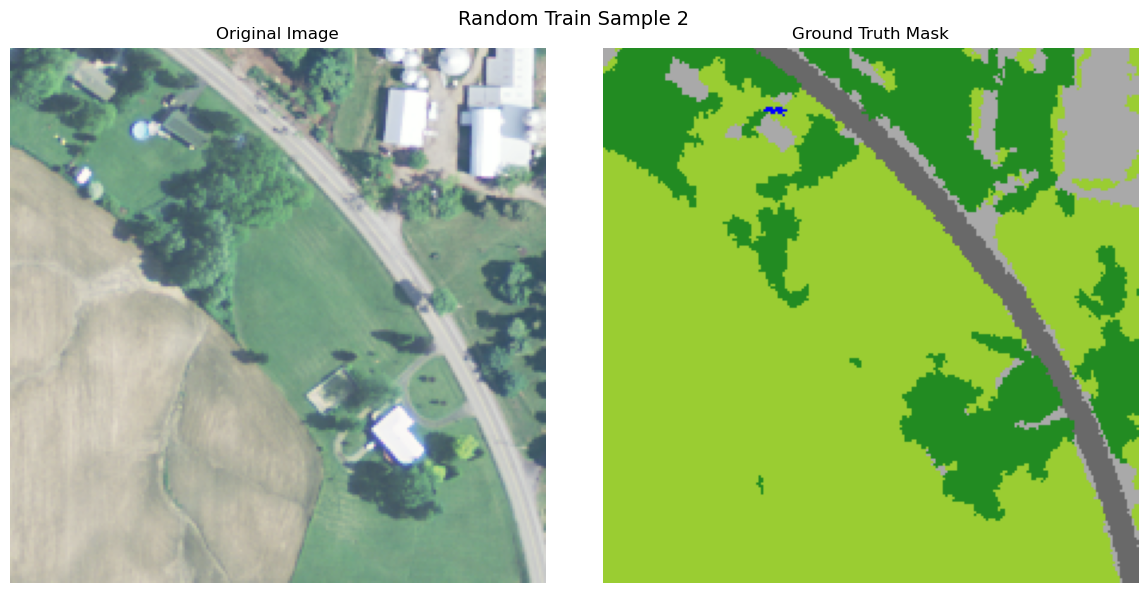
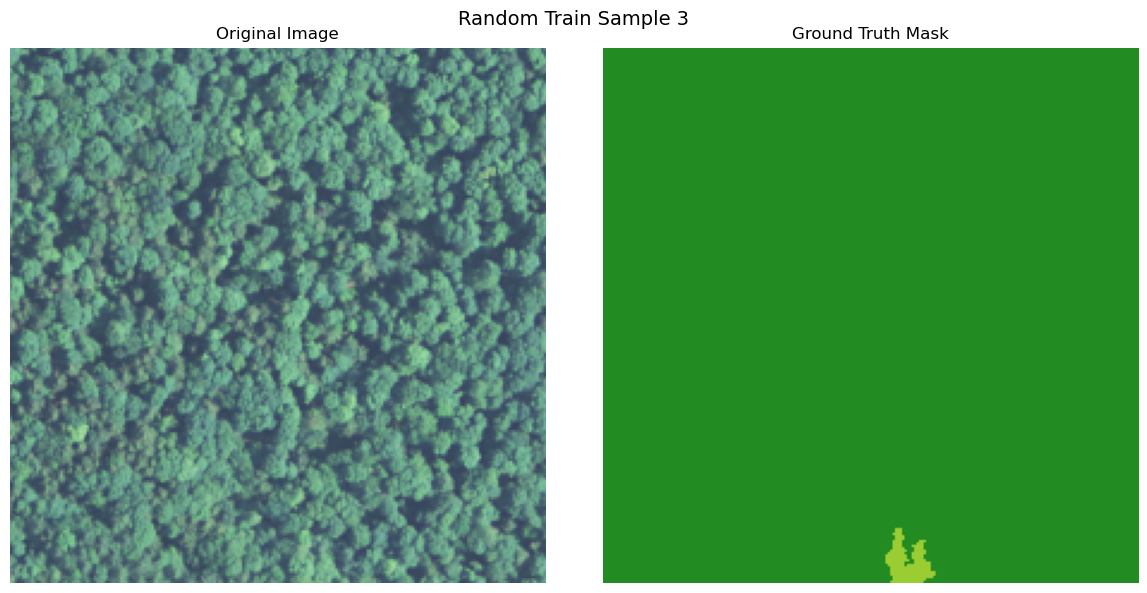
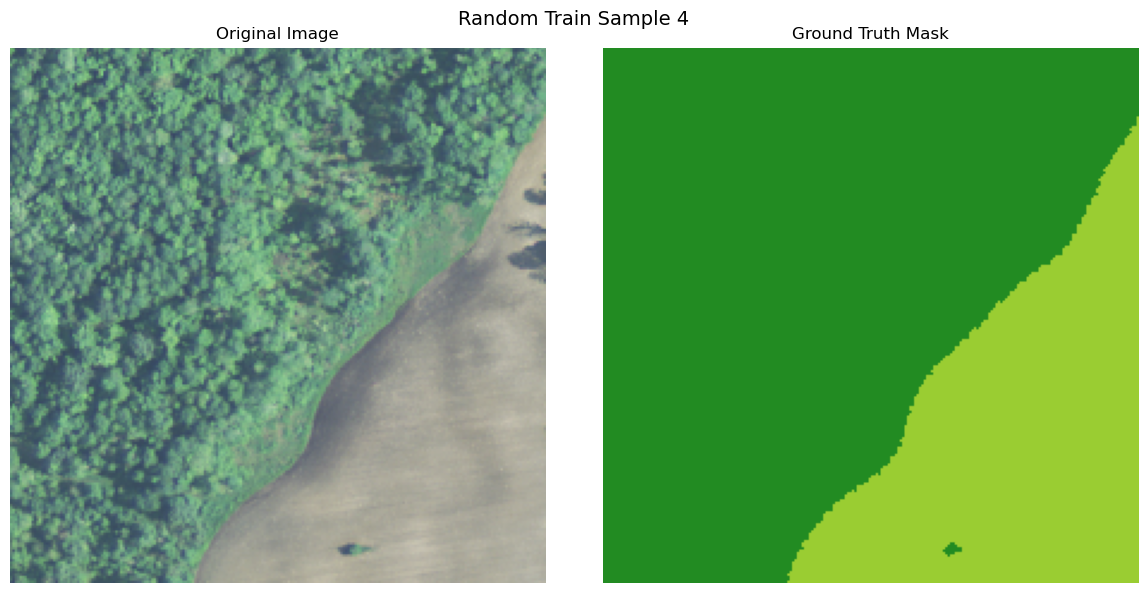
Uploading Data to Bluesight
Copy
import requests
from requests_toolbelt.multipart.encoder import MultipartEncoder
from pprint import pprint
def upload_file(file_path):
file_name = os.path.basename(file_path)
encoder = MultipartEncoder(
fields={'file': (file_name, open(file_path, 'rb'), 'application/octet-stream')}
)
response = requests.post(
f"{API_URL}/files",
data=encoder,
headers={'Content-Type': encoder.content_type}
)
if response.status_code == 200:
print("File uploaded successfully!")
return response.json()
else:
print(f"Failed to upload file. Status code: {response.status_code}")
print(f"Response: {response.text}")
return None
Copy
%%time
file_object = upload_file(output_file)
pprint(file_object, sort_dicts=False)
Copy
File uploaded successfully!
{'id': 'file-c6pyf79g',
'bytes': 262418408,
'created_at': 1722456495,
'filename': 'land_segmentation_train_data.h5'}
CPU times: user 1.5 s, sys: 298 ms, total: 1.8 s
Wall time: 2min 5s
Starting Training
Now we need to load data to the format which is accepted by our API. Essentially, it is raw pixels data from all bands with some metadata.Copy
def create_training_job(task, num_classes, training_file, validation_file=None):
payload = {
"task": task,
"training_file": training_file,
"validation_file": validation_file,
"hyperparameters": {"num_classes": num_classes}
}
response = requests.post(
f"{API_URL}/training/jobs",
json=payload,
headers={'Content-Type': "application/json"}
)
if response.status_code == 200:
print("Training job submitted successfully!")
return response.json()
else:
print(f"Failed to sumbit training job. Status code: {response.status_code}")
print(f"Response: {response.text}")
return None
Copy
training_job = create_training_job("segmentation", len(label_mapping), file_object["id"])
pprint(training_job, sort_dicts=False)
Copy
Training job submitted successfully!
{'task': 'segmentation',
'training_file': 'file-c6pyf79g',
'validation_file': None,
'hyperparameters': None,
'id': 'trainingjob-cjhzikag',
'created_at': 1722457908,
'status': 'initializing',
'error': None,
'trained_model': None,
'finished_at': None}
Copy
def retrieve_training_job(job):
response = requests.get(
f"{API_URL}/training/jobs/{job}"
)
if response.status_code == 200:
return response.json()
else:
print(f"Failed to retrieve training job. Status code: {response.status_code}")
print(f"Response: {response.text}")
return None
Copy
%%time
import time
status = None
while status not in ("succeeded", "failed", "cancelled"):
training_job = retrieve_training_job(training_job["id"])
status = training_job["status"]
print(status)
time.sleep(20)
pprint(training_job)
Copy
initializing
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
running
succeeded
{'created_at': 1722457908,
'error': None,
'finished_at': 1722459229,
'hyperparameters': None,
'id': 'trainingjob-cjhzikag',
'status': 'succeeded',
'task': 'segmentation',
'trained_model': 'model:segmentation-juo7qapo',
'training_file': 'file-c6pyf79g',
'validation_file': None}
CPU times: user 562 ms, sys: 52.9 ms, total: 615 ms
Wall time: 22min 29s
Run inference
For inference we need only images. You can check detailed endpoint specification in the docs.Copy
X_test = [{
"gsd": GSD,
"bands": BANDS,
"pixels": np.load(OUTPUT_DIR / "val/chips" / chip_name).tolist(),
"platform": COLLECTION
} for chip_name in tqdm(val_chip_names[:N_TEST_SAMPLES])]
y_test = np.array([np.vectorize(label_mapping.get)(np.load(OUTPUT_DIR / "val/labels" / chip_name).squeeze()).tolist() for chip_name in tqdm(val_label_names[:N_TEST_SAMPLES])])
Copy
100%|████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 100/100 [00:00<00:00, 116.81it/s]
100%|████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████████| 100/100 [00:00<00:00, 140.44it/s]
Copy
def run_trained_model_inference(model, images):
payload = {"model": model, "images": images}
response = requests.post(
f"{API_URL}/inference",
json=payload,
headers={'Content-Type': "application/json"}
)
if response.status_code == 200:
return response.json()
else:
print(f"Failed to run inference. Status code: {response.status_code}")
print(f"Response: {response.text}")
return None
Copy
%%time
y_pred = run_trained_model_inference(training_job["trained_model"], X_test)
y_pred = np.array(y_pred["labels"])
Copy
CPU times: user 3.79 s, sys: 183 ms, total: 3.97 s
Wall time: 1min 24s
Copy
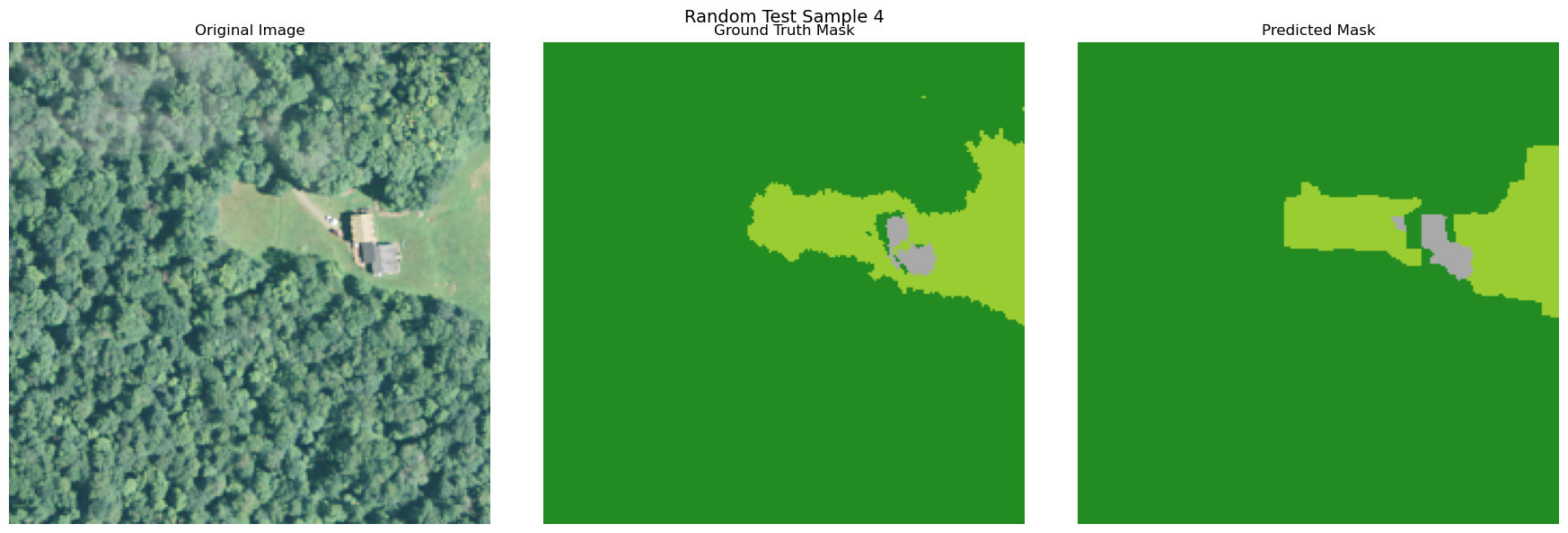
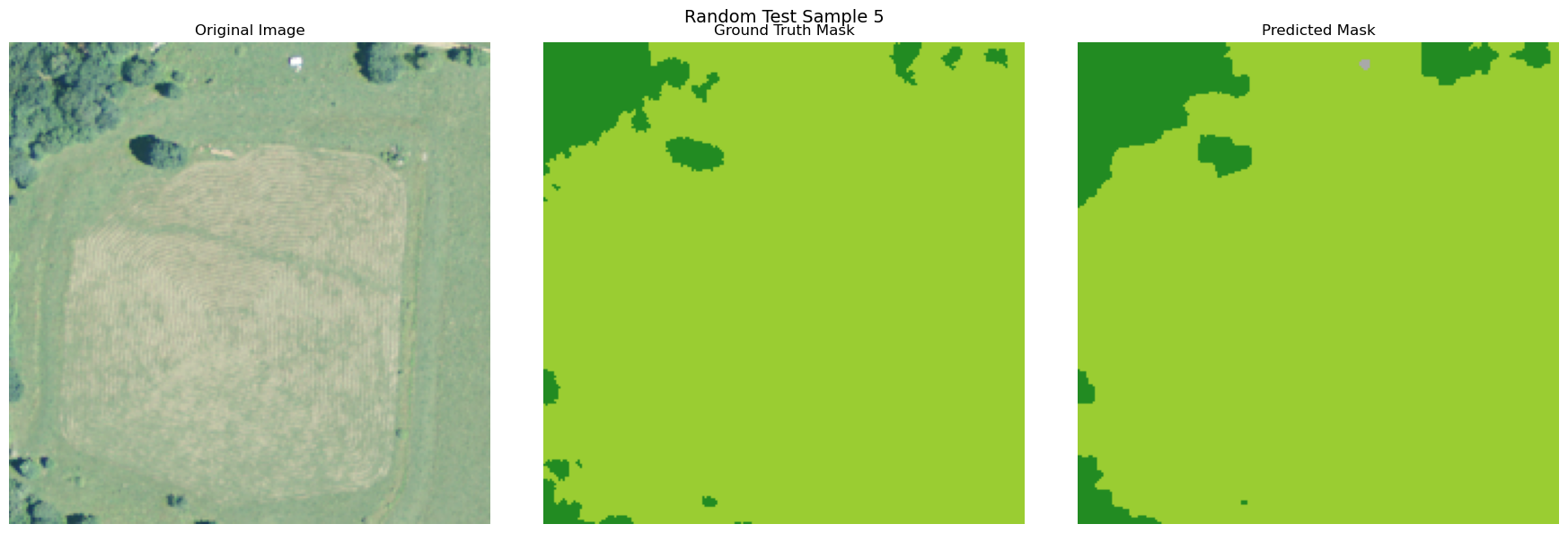
random_indices = np.random.choice(len(X_test), 5, replace=False)
for i, idx in enumerate(random_indices):
image = np.array(X_test[idx]['pixels']).transpose(1, 2, 0)
true_mask = y_test[idx]
pred_mask = y_pred[idx]
if image.shape[2] > 3:
image = image[:, :, :3]
visualize_image_mask_pair(image=image, true_mask=true_mask, pred_mask=pred_mask, title=f"Random Test Sample {i+1}")
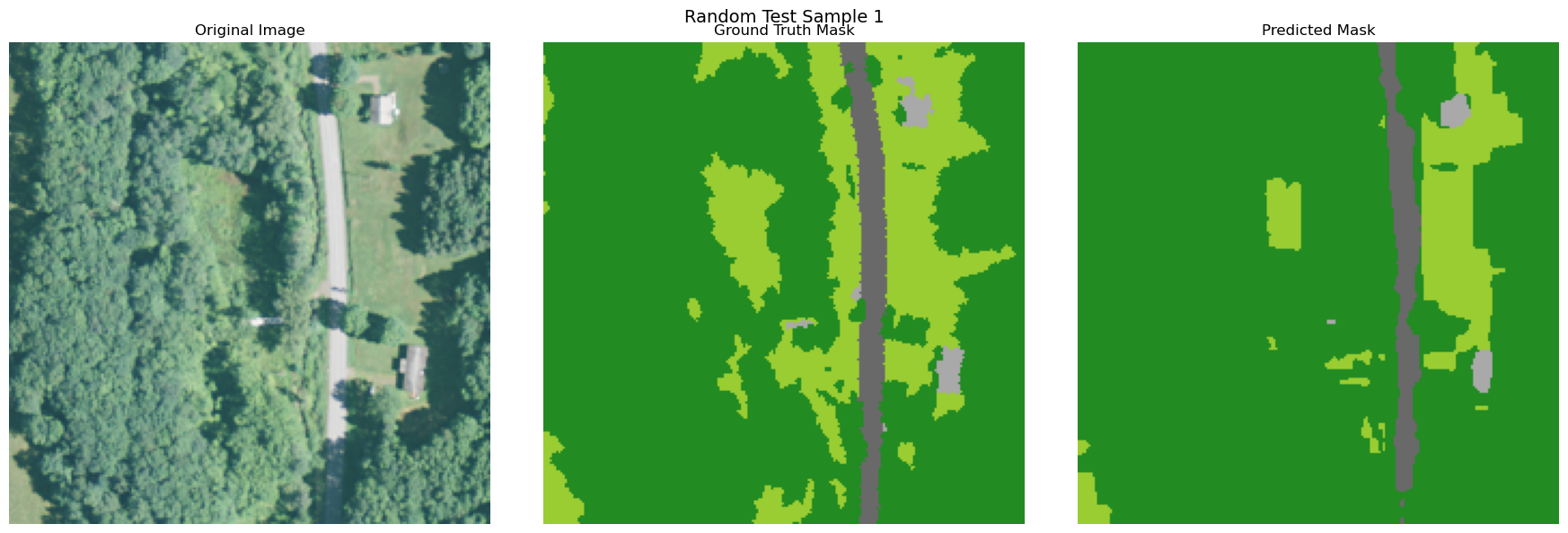
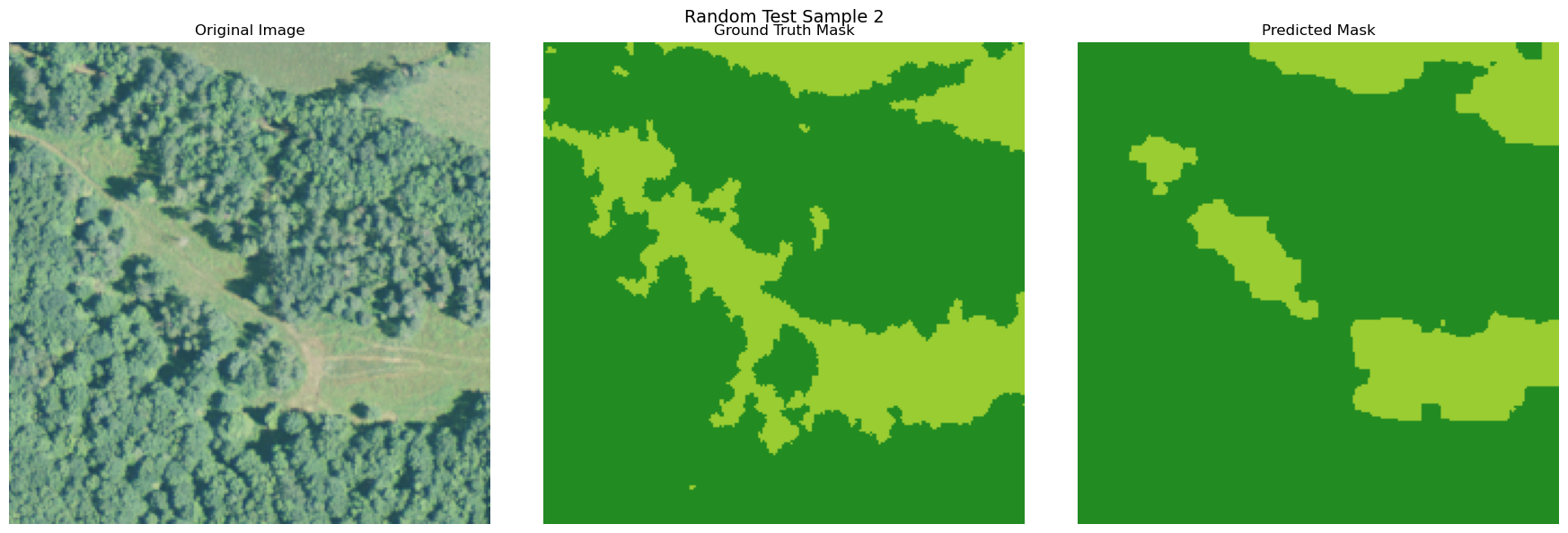
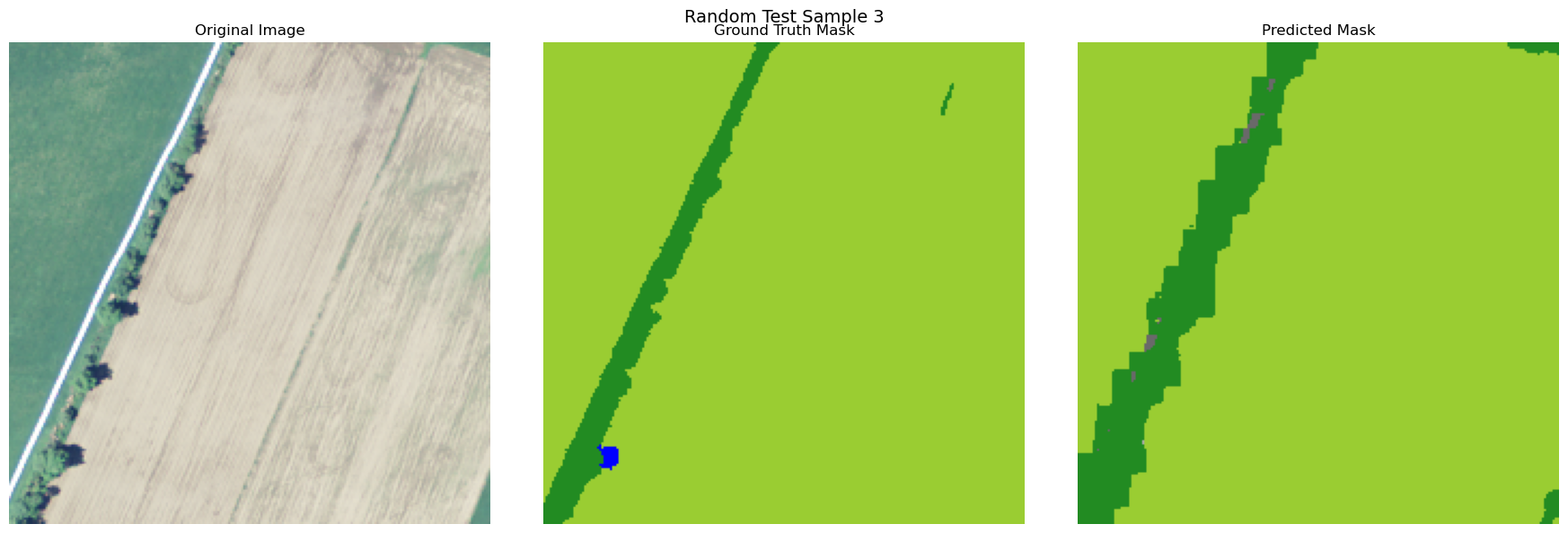
Copy
import numpy as np
from sklearn.metrics import confusion_matrix, accuracy_score, precision_score, recall_score, f1_score, jaccard_score
def calculate_segmentation_metrics(y_true, y_pred):
y_true_flat = y_true.reshape(-1)
y_pred_flat = y_pred.reshape(-1)
cm = confusion_matrix(y_true_flat, y_pred_flat)
pixel_accuracy = accuracy_score(y_true_flat, y_pred_flat)
mean_iou = jaccard_score(y_true_flat, y_pred_flat, average='macro')
precision = precision_score(y_true_flat, y_pred_flat, average='macro')
recall = recall_score(y_true_flat, y_pred_flat, average='macro')
f1 = f1_score(y_true_flat, y_pred_flat, average='macro')
return {
'confusion_matrix': cm,
'pixel_accuracy': pixel_accuracy,
'mean_iou': mean_iou,
'precision': precision,
'recall': recall,
'f1_score': f1
}
metrics = calculate_segmentation_metrics(y_test, y_pred)
for metric, value in metrics.items():
print(f"{metric}:")
print(value)
print()
Copy
/home/furiousteabag/.local/share/anaconda3/lib/python3.9/site-packages/sklearn/metrics/_classification.py:1318: UndefinedMetricWarning: Precision is ill-defined and being set to 0.0 in labels with no predicted samples. Use `zero_division` parameter to control this behavior.
_warn_prf(average, modifier, msg_start, len(result))
``````output
confusion_matrix:
[[ 141414 34499 1080 0 12725 802]
[ 28 2547949 34625 0 706 2085]
[ 223 353288 1724957 0 13209 3458]
[ 0 839 3413 0 27148 0]
[ 14 7318 1664 0 37640 4203]
[ 0 12461 4086 0 5970 41796]]
pixel_accuracy:
0.8955986926020408
mean_iou:
0.5495000999801362
precision:
0.6699292665323794
recall:
0.6568909938179078
f1_score:
0.6479898726350027
